Overview
Our research group is primarily interested in understanding the fundamental principles governing cellular protein homeostasis. The cellular environment must constantly adapt to changing conditions and one mechanistic strategy commonly employed is the targeted modification of proteins by ubiquitin which canonically leads to protein degradation by the proteasome. The ubiquitin proteasome system (UPS) serves as the cellular quality control system and it is tasked with the degradation of potentially toxic misfolded and misassembled proteins. The inability of the UPS to properly destroy these proteins has been linked to various human pathologies including amyotrophic lateral sclerosis (ALS, commonly referred to as Lou Gehrig’s disease), Parkinson’s disease, and many human cancers. The lab uses integrated molecular and systems-level approaches to understand ubiquitin-dependent regulatory mechanisms governing protein homeostasis. We are interested in how the proteome is dynamically remodeled in response to both chronic and transient stresses. Further, we want to understand how UPS function is altered during the progression of various human aging-associated disorders include neurodegenerative disorders and cancer.
As population longevity continues to increase, the incidence of aging-associated disorders such as cancer and neurodegenerative disease has also risen. Indeed the prevalence of neurological disorders has doubled in the past 20 years and continues to escalate such that it is estimated that over 100 million people worldwide will be diagnosed by 2050. Defects in the cellular pathways that limit the abundance or facilitate the removal of potentially toxic proteins have been linked to increased neurological dysfunction and decreased overall longevity in a variety of model organisms. Our goal is to develop new quantitative and global approaches to both monitor and manipulate protein homeostasis to alleviate aging-associated disorders.
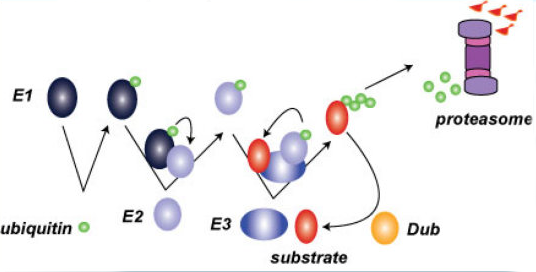
The decoding of mRNA into protein is a surprisingly error-prone process resulting in the continual production of defective translation products. Numerous defects in both the mRNA and the nascent chain can lead to translational stalling and subsequent degradation of both the defective translation product and the mRNA. Work in our lab seeks to understand how defective translation products are selectively targeted for degradation. Our goal is to define mechanisms that can be leveraged to both limit the production, and increase the clearance of defective translation products.
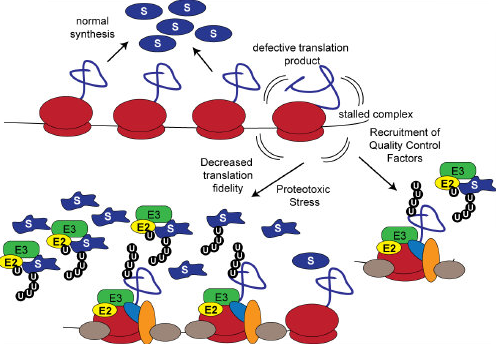
Resolving terminally stalled translational complexes
In response to limiting nutrient conditions, translation is reduced to prevent the continual production of proteins in a nutrient deficient cellular environment. Specific environmental stresses, such as oxidative or thermal stress, results in the sequestration of translation-competent mRNA into microscopically visible stress granules. Using a variety of proteomic and cellular approaches, our lab is investigating ubiquitin-dependent control mechanisms that govern the dynamics of mRNA sequestration into stress granules.
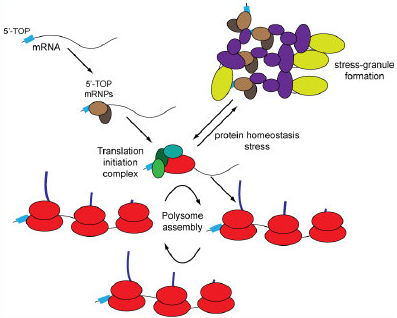
Stress Granule Dynamics
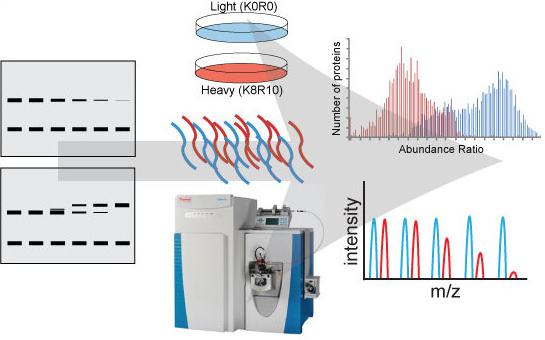
The cellular proteome is continually monitored by the protein folding and degradation machinery to prevent the accumulation of potentially toxic misfolded or misassembled proteins. Proteins that contain errors in their primary sequence that arise during the translation process need to be recognized and selectively degraded by the UPS. Improper function of protein homeostasis function has been hypothesized to be a contributing factor to a plethora of human degenerative diseases. We employ various proteomic approaches toward understanding global UPS function as well how this function is altered in response to stress. These approaches range from systematic analysis of protein networks associated with the UPS to a quantititive analysis of the ubiquitin-modified proteome.
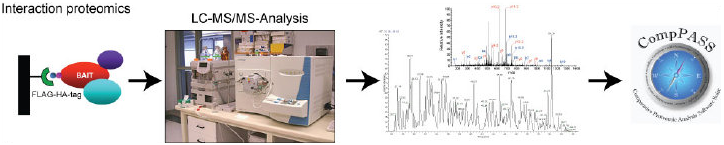
We are interested in trying to answer several questions related to quality control mechanisms. For instance, what are the ubiquitin ligases responsible for the ubiquitylation of mistranslated substrates? How do changes in free amino acid levels within the cell effect protein degradation and UPS function? Can we monitor alterations in UPS function in human pathologies associated with a decrease in quality control function? How does changing the redox potential within cells effect the quality control machinery? Does UPS function decline as we age? By understanding how proteins are recognized as quality control substrates and how the cell responds to protein homeostasis challenges we hope to develop strategies to modulate quality control function.
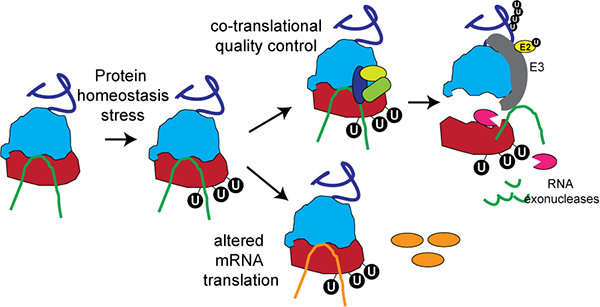
Ribosomal proteins represent some of the most abundantly ubiquitin-modified proteins in the cell. Work from our lab has demonstrated that activation of the unfolded protein response and inhibition of translation elongation induces site-specific ubiquitin modification of individual 40S ribosomal proteins. These ubiquitylation events appear regulatory in nature and do not target the protein for degradation. In fact, regulatory ubiquitylation of the ribosome is pervasive and largely unstudied. Current research is focused on understanding the mechanisms associated with catalyzing regulatory ribosomal ubiquitylation (RRub) as well as the underlying molecular function RRub imparts on protein homeostasis.